|
The INTERNATIONAL JOURNAL of APPLIED RESEARCH In Veterinary Medicine |
 |
| Current Issue |
| Previous Issues |
| Reprint Information |
| Back to The International Journal of Applied Research in Veterinary Medicine |
In utero Infection
of Pregnant Cattle by Mycobacterium avium Subspecies paratuberculosis
Detected by Nested Polymerase Chain Reaction
Claus. D. Buergelt, DVM, PhD
Elliot Williams, BS
Department of Pathobiology, College of Veterinary Medicine,
University of Florida, Gainesville, Florida
KEY WORDS: Bovine paratuberculosis, nested PCR, mycobacterial DNA, fluids and tissues, dam and fetus.
Abstract
This article describes
the applicability of a nested polymerase chain reaction (nPCR) using
2 sets of primers for the detection of Mycobacterium avium
subspecies paratuberculosis (Map) in infected fetal fluid and fetal tissues. The studies
were performed in 3 pregnant cows with clinical signs of paratuberculosis.
The cows were subjected to a complete necropsy. At necropsy, one of
3 fetuses of various gestational stages was positive for evidence of
Map when its fetal fluids and brain, liver, and lung were analyzed.
Its dam was positive for specific PCR reaction products in blood, milk,
ileocecal, mesenteric lymph nodes, and ileum. Two cows had negative
PCR results regarding fetal fluids and tissues, but positive PCR reaction
products in blood, milk, and tissues. The test system developed is simple,
cost-effective, and has a good turnaround time when compared with diagnostic
tests traditionally used for bovine paratuberculosis. The nested PCR
technique has a potential for application as an antemortem test to determine
in utero infection of fetuses in Map-infected cows through allantoi-aminocentesis.
Introduction
Paratuberculosis (Johne's disease), an insidious infectious disease of ruminants, occurs worldwide. The disease is caused by Mycobacterium avium subspecies paratuberculosis (Map), a facultative intracellular pathogen. Researchers have estimated that 5% to 20% of cattle in the United States are infected.1 Researchers currently believe that the principal pathway of transmission of Map is the fecal-oral route and that the calf is the most susceptible to such transmission.2 The fecal-oral transmission hypothesis has been used to formulate disease-control strategies for cattle. The isolation of Map from sites distant from the intestinal tract, the traditional targets for infection, such as the udder,3, fetus,4,5 kidney,6 liver,7 and male reproductive tract,8 have suggested active dissemination of Map and alternate transmission pathways such as milk, semen, and transplacental infection of fetuses. Such dissemination of Map should broaden the epidemiology concepts of paratuberculosis.
Culture studies have shown that between 20% and 40% of fetuses from cows with advanced Johne's disease were infected in utero, compared with 9% of fetuses being culture positive from asymptomatic infected cows.5 Because presently no way to identify such in utero-infected calves prenatally exists, the decision to keep a calf born to a proven-infected dam as replacement is if one parts with the dam difficult after infection detection regardless of the diagnostic method chosen. We report the feasibility of detecting infection in utero with a nested PCR technique developed in our laboratory for identifying Map in blood monocytes and milk macrophages and its potential application as prenatal test via placental fluid collection during pregnancy in the cow.
Materials and Methods
Animals
Three pregnant cows with clinical signs of Johne's disease such as weight loss and diarrhea were submitted for necropsy at the Veterinary Medicine Teaching Hospital to confirm the clinical diagnosis. The cows came from herds previously diagnosed to have Johne' disease. Cow 1 (N03-509) was a 5-year-old Angus 2 months into gestation. The cow was lactating and running a calf. Cow 2 (N03-541) was a 5-year-old Holstein, dry, and 6 months into gestation. Cow 3 was 4 years old, dry, and 7 months into gestation. A complete necropsy was performed on all 3 cows, including the fetuses, and target tissues were collected for histopathology and polymerase-chain reaction (PCR). Blood, milk when available, liver, spleen, mesenteric and ileocecal lymph nodes, ileum, liver, spleen were collected from the dams for PCR; placental fluids, abomasal contents, brain, lung, and liver tissue samples from the fetuses for PCR.
Specimen Handling
Blood of the dams was collected into EDTA-containing vacutainers via coccygeal vein bleeding after cleaning the tail skin with cotton soaked in alcohol. The blood samples were subjected to Ficoll-IsopaqueTM gradient centrifugation, and monocytes were harvested from the interface. When available, milk was collected into 50 mL sterile centrifuge tubes pooled from all 4 quarters after meticulous cleansing of the teat with cotton soaked in alcohol. The first drops of milk were voided. Milk was subjected to centrifugation at 1,000 g for 15 minutes. After centrifugation the supernatant was removed and discarded. The remaining samples were washed in PBS 3 times and suspended in 1 mL PBS for cell counting, resuspended in100 mL of O.2 N NaOH and boiled at 110°C for 20 minutes to extract crude DNA and centrifuged at 500 g for 3 minutes. Neutralization after NaOH extraction was not attempted. Tissues from mesenteric and ileocecal lymph nodes, liver, spleen, brain, lung, and ileum were prepared for PCR by touchpressing on glass slides. After air-drying, 200 mL of 0.2N NaOH were dispersed over tissues, and a sterile razor blade was used to scrape samples from the slides. Samples were placed into a 1.5-mL centrifuge tube and boiled at 110°C for 20 minutes. For the PCR reaction 1 mL of supernatant was chosen. Liquid samples from the allantois, amniotic, and fetal abomasum (25 mL) were centrifuged for 60 minutes at 1,000 g. Pellets were washed 3x with PBS, suspended in 0.2N NaOH and boiled as discussed previously.
PCR
A nested PCR was used. Briefly, for the first reaction primers P90, 91 were chosen, and primers J1, J2 were chosen for the second reaction. After DNA extraction, 1 mL of the lysates were submitted for PCR .A protocol of 35 cycles of 30 seconds at 94˚C, at 58˚C for 15 seconds and at 72˚C for 60 seconds was followed for the simple PCR. For the nested PCR, a program of 36 cycles with 30 seconds at 94˚C, 15 seconds at 63˚C and 60 seconds at 72˚C. A commercial reaction mix (Eppendorf Hotmaster Mix, Westbury, NY) was used according to the company's specifications. A volume of 10 mL of the PCR reaction products was run on 1.5% agarose gel by electrophoresis in TAE running buffer (Continental Lab Products, San Diego, CA). DNA extracted from Mycobacterium avium subspecies paratuberculosis laboratory strain No. 7283 was used as positive control for primers P90,91 and J1,J2 (nested control). Sterile water was used as negative control for the PCR assay. Gel inspection was performed using ultraviolet light and recorded with a computerized digital camera (UVP Transilluminator System, Upland, CA).
To avoid false-positive PCR results, tubes containing blood, milk, and fetal fluid samples were washed with alcohol before processing samples. Samples were initially processed in a sterile hood and then in 2 separated workstations, cleaned each time for each test with alcohol. Needles, syringe holders, and gloves were changed with each animal and for each step of the procedure.
Histopathology
Tissue sections were stained with hematoxylin-eosin (H&E) and acid fast stains (Fite's) and examined under a light microscope to qualitatively determine the extent of the granulomatous inflammation and number of acid-fast bacilli.
ELISA
The ELISA originally developed by W. D. Richards
(Allied Laboratories, Ames, IA) was performed with a crude, soluble
protoplasmic antigen (Allied Monitor, Fayette, MO). Test sera were preabsorbed
with Mycobacterium phlei.
Results were calculated from
wavelength readings (OD at 405 nm) of triplicates and recorded as negative
(< 1.5 OD), suspicious (1.5 to 1.9 OD), low positive (2.0 to 2.5
OD) and high positive (> 2.5 OD).
AGID
The same crude antigen
that was used for the ELISA was selected for the agar-gel immunodiffusion
test. The central well was loaded with 35 mL antigens. The peripheral
wells were inoculated with 45 mL of test sera.
A reference serum from a proven positive paratuberculous cow was used
as positive control. Final readings were performed after 48 hours.
Results
Necropsy confirmed that all 3 cows had Johne's disease, characterized by granulomatous enteritis and mesenteric lymphadenitis. Cow No. 1 (N03-509) had a female fetus measuring 22 cm crown to rump; cow No. 2 (N03-541) had a male fetus measuring 45 cm crown to rump; and cow No. 3 (N03-583) had a male fetus measuring 71 cm crown to rump. When target tissues of the dams were subjected to acid-fast stains, cows Nos. 1 and 2 showed evidence of paucibacillary Johne's disease; cow No. 3 was categorized pluribacillary.
Serologically, cows 1, 2, and 3 were positive on AGID. All 3 cows were positive on ELISA, with cow No. 1 showing strong positive (OD 4.3), cow No. 2 showing strong positive( OD 4.7), and cow No. 3 showing strong positive with OD's declining from 5.5 to 3.3 when performed consecutively every second day (7 times).
PCR reaction products from mesenteric lymph node, ileocecal lymph node, and ileum of cow No. 1 were positive with primers J1,J2, but negative with primers P90, P91. Single band signals were obtained at 333 bp. Blood and milk samples were positive on J1,J2, but not for P90,P91 (Fig. 1). Allantoic fluid, liver, lung, and brain from the fetus were positive with primers J1,J2 (Figs. 2, 3) at their respected molecular location, and negative with P90,P91. Amniotic fluid was not available for PCR analysis.
PCR reaction products from cow No. 2 were negative on blood when tested at time of euthanasia, but positive for J1,J2 on ileum, cecum, mesenteric, and ileal lymph nodes and liver, but not on spleen; no signals were found when tested with primers P90,P91. No PCR reaction products were found with the first and second primer sets, when fetal allantoic, amniotic, abomasal fluids, and tissues from lung, brain and liver were tested.
Cow No. 3 (N03-583) was subjected to repeat serologic and blood PCR testing before euthanasia was performed. Blood PCR reaction was positive on J1,J2 on 4 of 7 occasions when samples were obtained every second day. Blood was also positive on one occasion when tested with primers P90,P91. Milk was not available because the animal was dry. Tissues collected from the dam at postmortem were positive for Map DNA in ileocecal and mesenteric lymph nodes, ileum, and jejunum when subjected to J1,J2 primers, but negative for P90,P91. Liver and spleen were negative to both sets of primers. Fetal plancental fluids, abomasal fluids, and all fetal tissues were negative to both sets of primers.
Discussion
The detection of Map DNA by PCR in fetal fluids and various fetal tissues supports the concept of Johne's disease being a disseminated infectious disease and should draw attention to avenues of transmission other the traditional fecal-oral pathway of transmission. Extraintestinal pathways such as milk and in utero modes should be addressed when considering the epidemiology and control of the disease through testing and culling. The management often considers economic hardships over medical recommendation to remove infected dams and their offspring. The diagnosis of prenatal infection via fetal fluid analysis may give solid scientific evidence for the correctness of such recommendation over predicted values through calculations of statistic likelihoods from the literature.
Application of bovine fetal fluid collection is a relatively unexplored area in veterinary medicine. It has been sporadically reported in the literature and exclusively for the purpose of determining the gender of the unborn calf through PCR.9,10 In these instances, fetal fluid was recovered via ultrasound-guided aspiration, but because of technical difficulties often resulted in fetal death through maternal contamination at the site of the vagina.9,10 The volume of fluid recovered with such technique ranged from 0.5 to 5 mL in cows 61 to 120 days pregnant.11 This may not be enough volume for the nested PCR perceived for the determination of infection status at that age of gestation. Volume of fetal fluid increases with gestational age. The volume of amniotic fluid increases up to 2.5 L in the seventh month of pregnancy, and that of the allantois to 9 L.11 The in utero application of antibiotics may help to minimize the risk of fetal death after transvaginal puncture technique for fetal fluid collection. The development of safer collection techniques would be of benefit to the concept of in utero determination of Map infection without risk to the developing fetus.12,13 Safer collection techniques in combination with molecular diagnostic techniques promise to have prediction potential for the decision of removing the infected dam and her offspring from the herd. Culling the most recent calf from a cow being diagnosed as infected may not be necessary in some instances when prenatal diagnosis is negative for Map.
Cultural isolation of Map from fetuses has been reported on several occasions from the UK and US.4 From a small collection of pregnant infected cows sent to slaughter, 4 of 9 fetuses of all gestational periods were culture positive on Herrold's egg yolk medium (Buergelt, PhD Thesis, Ithaca, NY, 1976). In 3 of the fetuses, spleen, kidneys, and placenta were positive on culture. Liver, lung, and intestine contained Map organisms in 2 fetuses. The bacilli were isolated from fetal brain, gastric contents, and amniotic fluid in one instance. A tissue reaction such as inflammation to the presence of organisms was not appreciated microscopically in all fetuses.
Epidemiologic observations of a few cows turning into clinical cases of Johne's disease at a young age such as 2 years and before the first calving have led to the hypothesis that these animals might have been exposed to in utero infection when compared with the usually 3- to 5-year-old animals that are expected to have been infected postnatally.4
Our results identified one out of three fetuses from clinical cows by PCR to be infected with Map. This small number corroborates with previous reports showing by culture that 20% to 40% of fetuses are infected in utero.5
PCR is a tool to amplify DNA specific to mammalian and prokaryotic cells. In case of M. avium subspecies paratuberculosis (Map), the claimed species-specific insertion sequence IS 900 is probed for with primers P90,P91 to yield a reaction product of 413 bp.14 The second set of primers, J1,J2, was designed to be used in a nested PCR assay. It spans a 333-bp region within the P90,P91 region. PCR assays for IS 900 have been reported to be able to detect as low as 104 colony-forming units per gram of feces in shedding cows.15 A nested PCR allows further amplification of the signal already amplified by the first pair of primers, increasing the specificity and sensitivity of the test system.
The developed nested PCR appears to have a potential to discover Map DNA in blood and milk phagocytes of clinical and also silently infected cattle. It is field-applicable, cost effective, and has a turnaround time of 8 hours for individual blood, milk, fetal fluid, and tissue samples. Larger numbers of samples can be handled within 48 hours. The test appears to have promise to identify subclinical animals before ELISA or AGID tests become positive and before fecal culture is successful as concluded from a small series of 56 animals (study to be published in the Australian Journal of Veterinary Medicine). In terms of predicting the presence or absence of in utero infection, the nested PCR test may have a place as an antemortem test in the future after safer and more user-friendly clinical fetal fluid collection techniques are developed.
Acknowledgments
We would like to thank Drs. A. Donovan, O. Rae, and C. Risco, from the Department of Large Animal Clinical Sciences, University of Florida, for providing us with the pregnant cows.
References
1. Thoen CO, Braun KH: Current knowledge on paratuberculosis. J Am Vet Med Assoc 192:1609-1611,1988.
2. Corner AL, Abbot KA, Pfeiffer DU: Unanswered questions about transmission of Mycobacterium avium subsp paratuberuclosis. Vet J 165:182-183, 2003.
3. Sweeney RW, Whitlock RH, Rosenberger AE: Mycobacterium paratuberculosis cultured from milk and supramammary lymph nodes of infected asymptomatic cows. J Clin Microbiol 30:166-177, 1992.
4. Sweeney
EW: Transmission of paratuberculosis. Vet Clin North Am
Food Anim Pract 12:305-312,
1996.
5. Sweeney RW, Whitlock RH, Rosenberger AE: Mycobacterium paratuberculosis isolated from fetuses of infected cows not manifesting signs of the disease. Am J Vet Res 53:477-480, 1992.
6. Hines SA, Buergelt CD, Wilson JH, et al: Disseminated Mycobacterium paratuberculosis in a cow. J Am Vet Med Assoc 190:681, 1987.
7. Barrington GM, Gay JM, Eriks IS et al: Temporal patterns of diagnostic results in serial samples from cattle with advanced paratuberculosis. J Vet Diagn Invest 15:195-200, 2003.
8. Larsen AB, Kopecky KE: Mycobacterium paratuberculosis in reproductive organs and semen of bulls. Am J Vet Med Assoc 31:255-258, 1970.
9. Vos PLAM, Pieterse MC, van der Weyden MAM, et al: Bovine fetal fluid collection: transvaginal, ultra-sound guided puncture technique. Vet Rec 127:542-546, 1999.
10. Makondo K, Amiridis IA, Jeffcoate PJ, et al: Use of polymerase chain reaction to sex the bovine fetus using cells recovered by ultrasound-guided fetal fluid aspiration. Ann Reprod Sci 49:129-133, 1997.
11. Wintour EM, Laurence BM, Lingwood BE: Anatomy, physiology and pathology of the amniotic and allantoic compartments in the sheep and cow. Aust Vet J 63:216-224, 1980.
12. Sprecher DJ, Kaneene JB: Diagnostic techniques for transvaginal transuterine aspiration of bovine fetal fluid during early fetal period. Theriogenology 38:581-587, 1992.
13. Garcia A, Salaheddine M: Bovine ultrasound-guided transvaginal amniocentesis. Theriogenology 47:1003-1008, 1997.
14. Vary PH, Andersen PR, Green E, et al. Use of highly specific DNA probe and polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne's disease. J Clin Microbiol 28:933-937, 1990.
15. Wipple DL, Kapke, PA, Andersen PR: Comparison
of a commercial DNA probe test and the cultivation procedures for detection
of Mycobacterium
paratuberculosis in bovine feces. J Vet Deign Invest 4:23-27, 1992.
Figure 1. Gel electrophoresis of J1,J2 amplification
products of blood, milk, and allantoic fluid from cow-fetus No. 1 (N03-509).
Lane 1: positive DNA control from lab strain No. 295; lane 2: milk;
lane 3: blood; lane 4: allantoic fluid sediment; lane 5: allantoic fluid
supernatant; lanes 6 and 7: negative control; lane 8: void; M: molecular
markers.
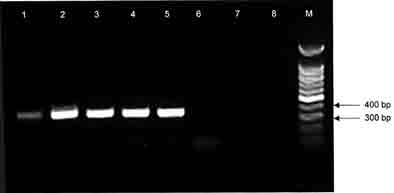
Figure 2. Gel electrophoresis of J1,J2 amplification products in allantoic fluid (N03-509). Lanes 1 and 2: positive DNA controls from laboratory strain No. 295; lane 3: allantoic fluid; lanes 4 and 5: negative controls; lane 6: molecular markers.
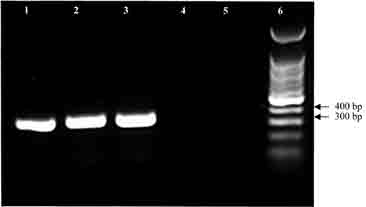
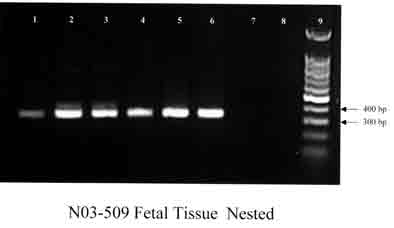
Figure 3. Gel electrophoresis of J1,J2 amplification
products of fetal tissues (N03-509).
Lanes 1 and 2: positive DNA controls from laboratory strain No. 295;
lane 3: liver; lane 4: lung; lane 5: brain; lane 6: spleen; lanes 7
and 8: negative controls; lane 9: molecular markers.
ISSN# 1542-2666